Principles of cell fate specification
In the Askary lab, we strive to find simple principles that are used during embryonic development to make complex tissues. To this aim, we develop innovative tools to monitor, characterize, and manipulate cells. We leverage both our methods and our findings to make better therapeutic strategies and molecular diagnostics.
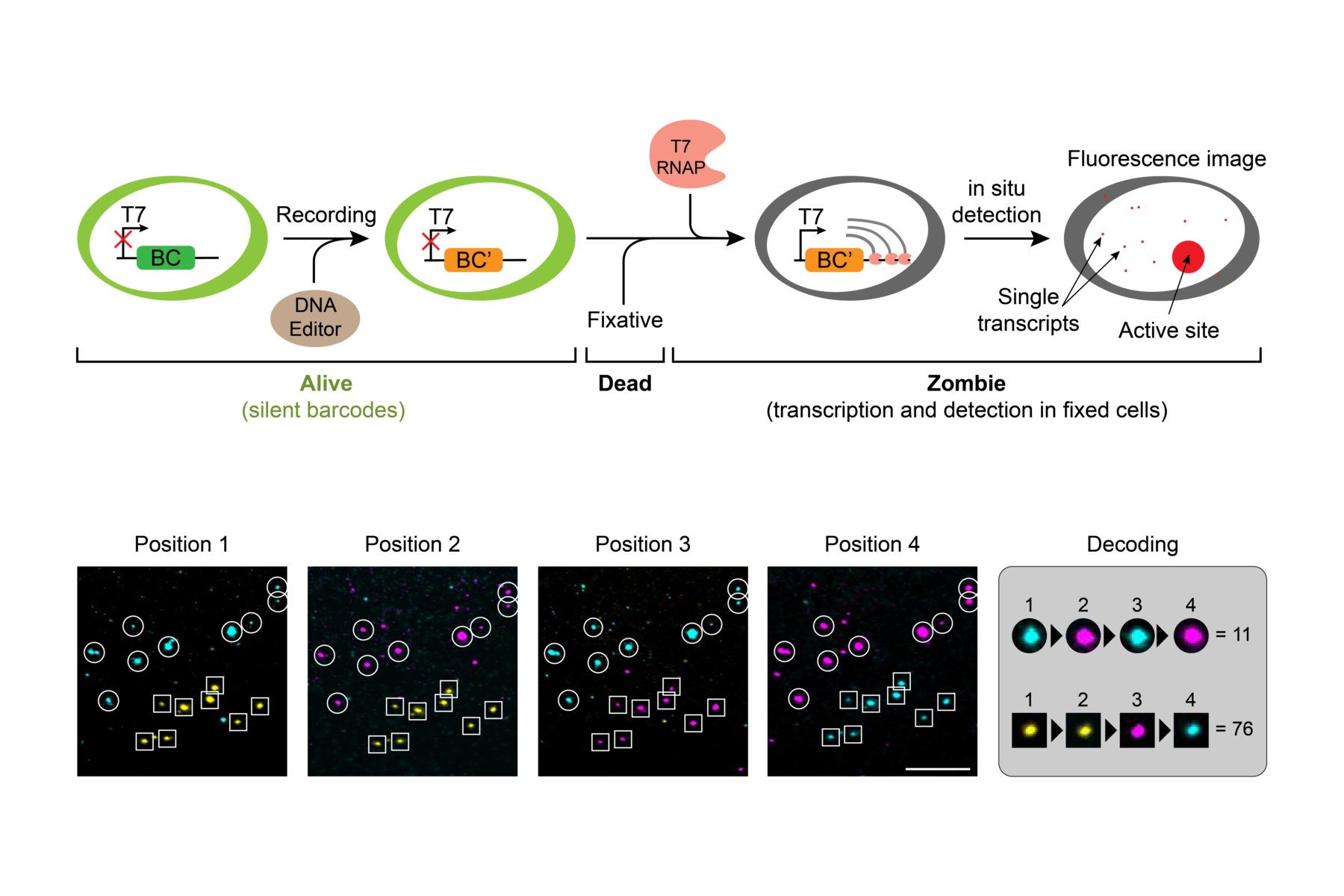
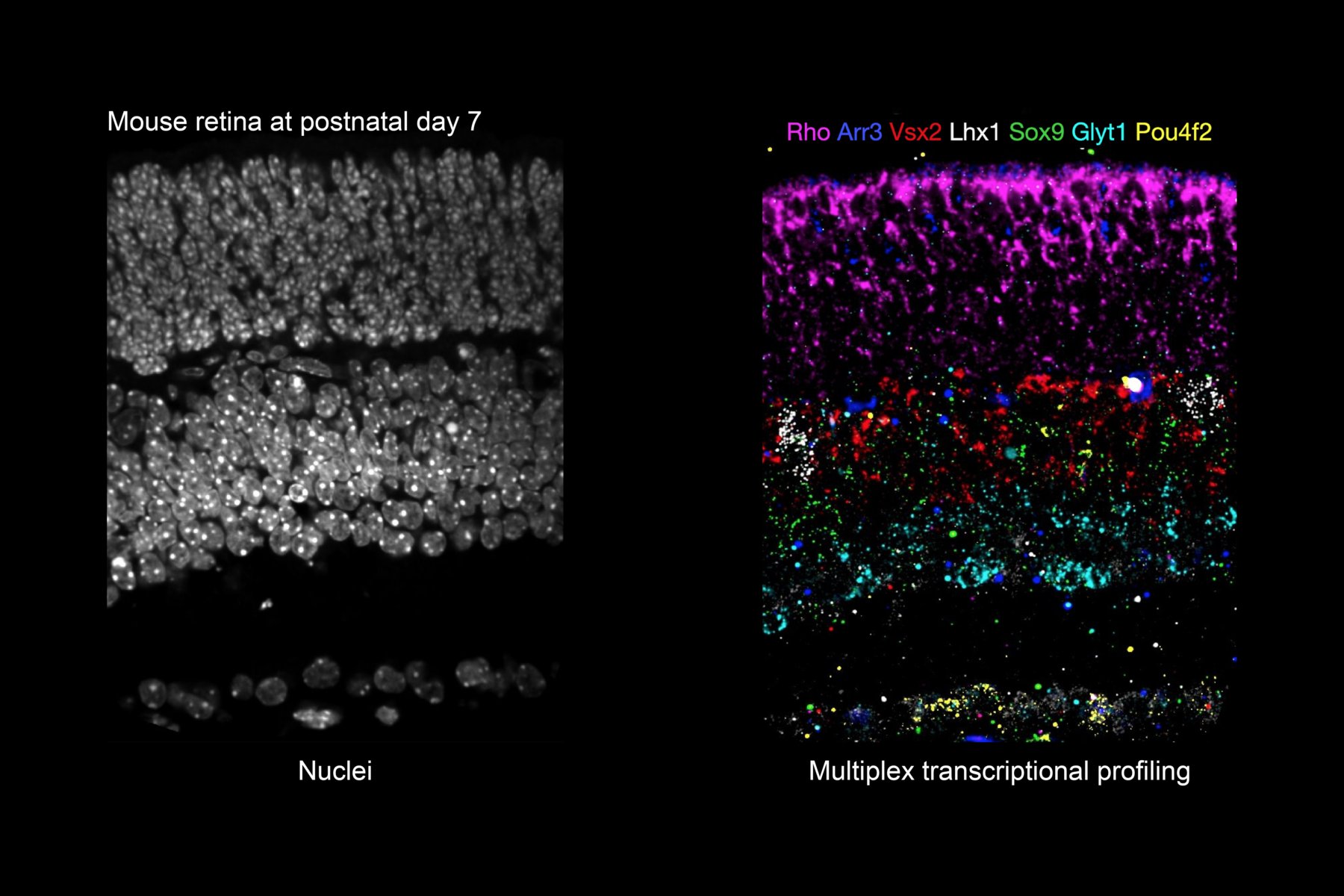
Identity and pattern of the cells are influenced by their intrinsic states, extrinsic signals, and stochastic processes. Collecting quantitative data is the first step to understand how these factors work together to make a functional tissue and how they go wrong in developmental and degenerative disorders. We have developed a method for sensitive and scalable in situ readout of DNA barcodes (Askary et al., 2020 Nature Biotechnology). Now our team is using this technology, together with CRISPR base editors, to monitor the intensity and order of signals that cells receive over time. We also use spatial transcriptomics to map gene expression across developing tissues, including retinal progenitor cells, early stage mouse embryos, and stem cell derived embryo models. To find interesting patterns in these data, that can lead us to principles of cell fate determination, we use computational tools of systems biology. We believe new technologies and interdisciplinary approaches are essential for providing new insight into the oldest questions in developmental biology.